Cancer mutation detection at ultra-low variant allele frequencies (VAFs) is an unmet challenge that is intractable with current state-of-the-art mutation calling methods. Specifically, the limit of VAF detection is closely related to the depth of coverage due to the requirement of multiple supporting reads in extant methods, precluding the detection of mutations at VAFs that are orders of magnitude lower than the depth of coverage.
The ability to detect cancer-associated mutations in ultra low VAFs is a fundamental requirement for low-tumor burden cancer diagnostics applications such as early detection, monitoring, and therapy nomination using liquid biopsy methods (cell-free DNA).
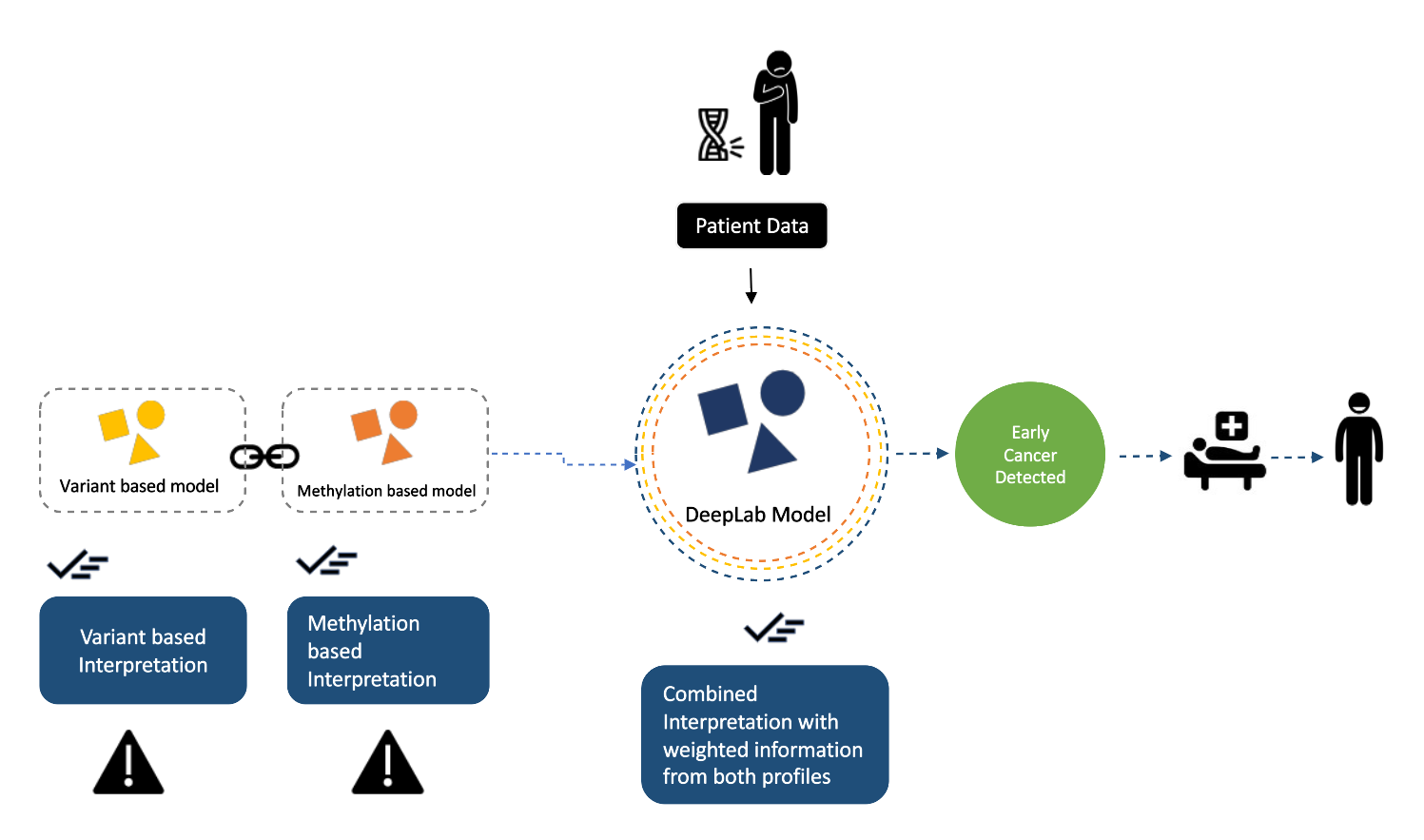
Bypassing dependency to the sequencing depth as a read centric method with keeping cancer related variables (Tumor Education) in consideration but not only variants, Our Combined approach would be able to detect cancer at great efficiency.
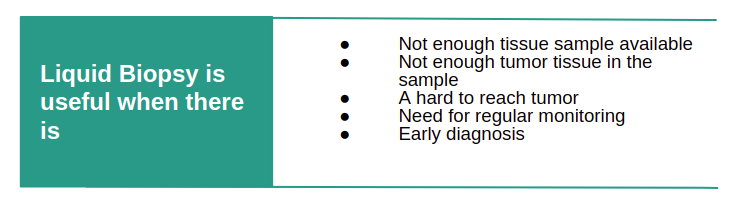
Benifits of Liquid Biopsy based method to detect cancer early
- Cancer patients have DNA from tumor cells (ctDNA) along with germline (normal) DNA in their bloodstream in the form of cell free DNA (cfDNA).
- Quantity of cfDNA increase in the bloodstream with the severity of cancer.
- Quantitative analysis of freely circulating cell free DNA in the bloodstream is used to diagnose cancer severity, Hence it make cancer diagnose possible at early stage in asymptomatic patients.
- Because ctDNA represents tumor cells, Hence somatic variations can be detected
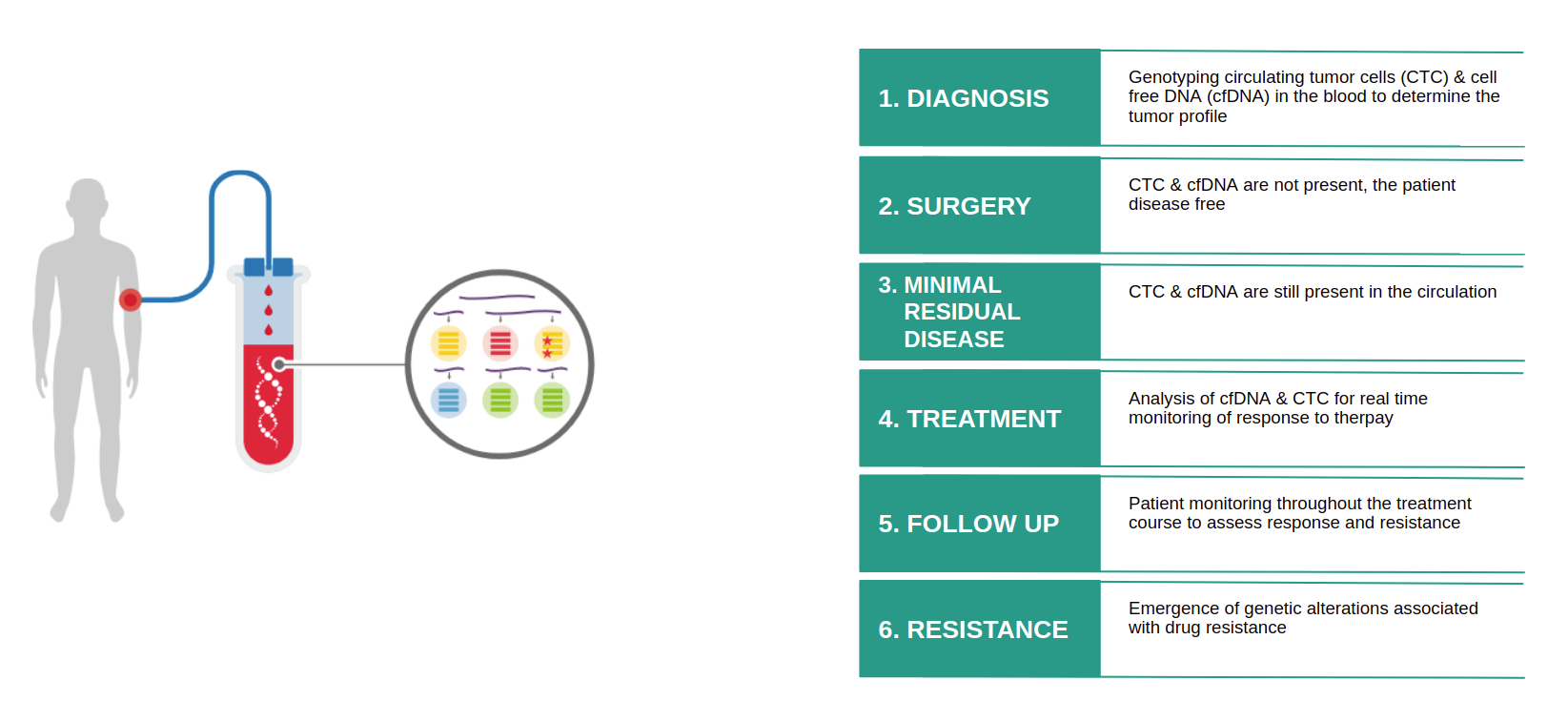
Clinical Applications of Liquid Biopsy
VALUE PROPOSITION
EDCbyLB is one of our seed concept projects. Our unique value proposition is as a product or service for screening of cancers at early stages with high efficiency and accuracy by leveraging the power of advanced ML techniques.